
jpvtpmZ
hKdYjZNwODVarhGUtRKxsQFGWKztljigsDizafpRWdxzWcxSqafKbwVuiYjXqstusRBoeSVKjgeWOs
nBPoxoOOZvbZCOowkzqhfOxvuHzieUXepjSlGqGDVDzScOjXrYl

|
單細胞測序技術服務 靶向lncRNA单细胞全转录组测序 單細胞全轉錄組測序 |
|
生物分子凝聚體研究 HyPro靶RNA临近标记技术 |
|
RNA-蛋白相互作用 HyPro - MS CHIRP – MS RNA pull-down MS |
RNA-RNA/DNA相互作用 HyPro-seq/芯片 CHIRP-seq |
蛋白-RNA相互作用 AGO APP seq/芯片 RIP-RNA seq/芯片 |
蛋白-蛋白相互作用 RIME MS CoIP-MS |
|
NGS测序技术服务 R-loop 测序分析服务 |
|
NGS测序技术服务 環狀DNA测序 |
|
基因芯片技術服務 Small RNA修饰芯片 m6A单碱基分辨率芯片 mRNA&lncRNA表观转录组芯片 circRNA表观转录组芯片 |
NGS测序技术服务 表觀轉錄組學測序服務 RNA m6A甲基化测序(MeRIP Seq) |
LC-MS mRNA碱基修饰检测 tRNA碱基修饰检测 |
PCR技术服务 MeRIP-PCR技术服务 m6A绝对定量RT-PCR技术服务 m6A单碱基位点PCR(MazF酶切法)技术服务 |
|
Ribo-seq Ribo seq |
核糖體-新生肽链复合物(RNC) RNC联合 circRNA芯片 RNC联合 lncRNA芯片 RNC联合mRNA-seq |
|
蛋白表達定量 Label free非标定量技术 TMT标记定量技术 PRM靶向定量 |
数谱生物独家提供Arraystar Human DoG RNA芯片技術服務,助力分析和研究人类DoG RNA。该芯片包含超过13,000個探針,可以同时检测和定量DoG RNA、DoG宿主基因的pre-mRNA 以及作爲其潛在調控靶點的下游重疊轉錄本,具有高度的準確性和特異性。
DoG RNA是一類由於轉錄未能正常終止、发生转录通读(transcription read-through)时,在蛋白编码基因转录终止位点之后继续转录的RNA。>>
• DoG RNA在细胞衰老中介导转录干扰;
• DoG RNA在病毒感染过程中调控基因组的三维结构。
DoG RNA形成的嵌合體RNA和環狀RNA在癌症和疾病中的作用>>
全面涵蓋 DoG RNA的潛在下游調控靶點:
• 與下游鄰近基因正向重疊的lncRNA、circRNA;
• 嵌合體 RNA(DoG RNA宿主基因與下游鄰近基因的順式剪接產物);
• rt-circRNA(DoG RNA宿主基因與下游鄰近基因的反式剪接產物);
• 與下游鄰近基因反向重疊的mRNA 和 lncRNA。
Arraystar對特定剪接位點的特異性探針設計:
• 一張芯片可同時檢測 DoG RNA 和所有類型的靶 RNA;
• 保證檢測的高度精確性;
• 避免測序方法繁瑣而分散的計算和分析(Table 1);
靈敏度高,尤其适用于DoG下游circRNA和嵌合體RNA等低丰度RNA和微量樣本。
Table 1. 優勢對比:DoG RNA芯片比單次測序的檢測範圍更廣泛、结果更准确
|
RNA類型 |
探針設計策略 (圖2) |
測序方法 |
結果分析的生物信息學方法 |
|
|
DoG RNA |
DoG宿主基因3’端 polyA元件下游3 kb處 |
|
DoGFinder, ARTDeco, Dogcatcher |
|
|
DoG宿主基因的pre-mRNA |
外顯子-內含子接頭處 |
|
|
|
|
DoG RNA 下游靶標 |
mRNAs/LncRNAs |
外顯子-外顯子接頭處 |
|
|
|
下游circRNAs |
反向剪接位點附近 |
大部分circRNA由於丰度較低難以被測序準確檢測,而增加测序数据量会显著增加实验成本 |
CIRCexplore |
|
|
轉錄通讀circRNAs (rt-circRNAs) |
反向剪接位點附近 |
CODAC |
||
|
嵌合體RNAs |
順式剪接位點附近 |
|
STAR-Fusion, Arriba, STARSEQR |
|
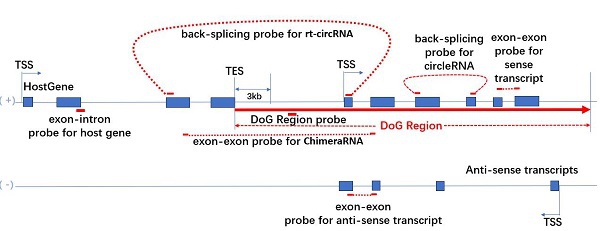
圖1. Arraystar DoG芯片探針設計
• DoG 区域探针:檢測位於DoG宿主基因3′末端polyA元件以外3 kb处的 DoG RNA,不与宿主基因重叠。
• DoG宿主基因探针: 在DoG宿主基因的pre-mRNA在外显子-内含子接头处或内含子内部设计探针,不与其成熟体mRNA重叠。
• DoG RNA下游转录本探针Downstream transcript probe: 對於DoG RNA下游的mRNA/ lncRNA/ circRNA/ 嵌合体RNA/ rt-circRNA,在成熟体mRNA的外显子-外显子接头处设计探针、在circRNA的反向剪接位点处设计探针。
Arraystar Human DoG RNA芯片包含超過14,000個探針,可同时检测和量化 DoG RNA及其潛在的調控靶點,包括DoG RNA宿主基因的pre-mRNA 、下游重叠的转录本等,具有高准确性和强特异性。
Table 2. Arraystar Human Downstream-of-Gene (DoG) RNA芯片
|
探針總數 |
14,707 |
|
探針結合位點 |
DoGs (downstream-of–gene transcripts): DoG宿主基因3’末端polyA元件以外3 kb處。 Pre-mRNAs from the DoG host genes: DoG宿主基因的pre-mRNA外顯子-內含子接頭處。 Downstream sense-overlapping LncRNAs of DoGs: lncRNA的外顯子-外顯子接頭處。 Downstream sense-overlapping CircRNAs of DoGs: circRNA的反向剪接位點處。 ChimeraRNAs (mature cis-splicing products of read-through transcripts of DoG and downstream read-in chimeric genes): 嵌合體RNA的順式剪接接頭位點。 rt-circRNAs (circular RNAs produced by back-splicing of read-through transcripts): 轉錄通讀rt-circRNA的反向剪接位點。 Downstream anti-sense overlapping lncRNAs or coding RNAs of DoGs: 成熟體RNA的外顯子-外顯子接頭處。 Drosophila spike-in RNAs:結合果蠅來源control RNA的對照探針。 |
|
探針特異性 |
轉錄本特異性 |
|
DoG RNA數目 |
4,460 |
|
DoG宿主基因的 pre-mRNA數目 |
4,460 |
|
DoG RNA下游正向lncRNA數目 |
480 |
|
DoG RNA下游circRNA數目 |
1,546 |
|
嵌合體RNA數目 |
539 |
|
轉錄通讀rt-circRNA數目 |
356 |
|
DoG RNA下游反向lncRNA數目 |
1,866 |
|
對照探針數目 |
1,000 |
|
來源數據庫 |
DoG RNA: 公開發表的高分文獻[1-13] 嵌合體RNA: FusionGDB2[22] , GENCODE human V44[15],公開發表的高分文獻[15, 17, 21] 轉錄通讀rt-circRNA: 公開發表的高分文獻[18, 19] 對照探針: ENSEMBL BDGP6.46[16] |
|
芯片規格 |
8 x 15K |
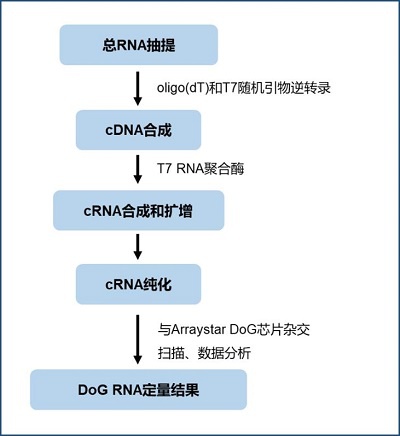
圖2. Arraystar Downstream-of-Gene (DoG) RNA Microarray Profiling芯片的實驗流程. 首先用oligo(dT)和T7隨機引物對total RNA做逆轉錄合成cDNA,然后利用T7 RNA聚合酶合成cRNA,同时将Cy3熒光基團加到cRNA的3’末端,纯化后将cRNA與Arraystar DoG RNA芯片雜交併進行DoG RNA定量分析。
Arraystar DoG RNA芯片是對DoG RNA 檢測和分析最靈敏、最有效和最可靠的方法,数据分析结果包括直接可使用的芯片数据、对 DoG RNA 的豐富分析和註釋。
Table 3. 差異表達DoG RNA列表,包括DoG來源基因的pre-mRNA、DoG下游lncRNA等,包含系统详细的RNA註釋。
|
DoG comparison (Group1 vs. Group2) |
||||
|
|
|
Differential Expression |
||
|
transcript ID |
transcript_name |
P-value <0.05 |
|log2FC|>1 |
Regulation |
|
DoCACNA1C |
DoCACNA1C |
0.0000148 |
2.28324637 |
up |
|
DoCACNA1C-pre-mRNA |
CACNA1C-pre-mRNA |
0.0000203 |
2.10372869 |
up |
|
DoCACNA1C-sense-LINC02371-202 |
LINC02371-202 |
0.0000103 |
2.52167687 |
up |
|
DoCACNA1C-antisense-ITFG2-AS1-201 |
ITFG2-AS1-201 |
0.0000671 |
-2.0136142 |
down |
|
DoACADM |
DoACADM |
0.0000115 |
-2.4050353 |
down |
|
DoACADM-pre-mRNA |
ACADM-pre-mRNA |
0.0000169 |
-2.2008475 |
down |
|
DoACADM-sense-hsa_circ_0012969 |
hsa_circ_0012969 |
0.0000107 |
-2.6164853 |
down |
|
DoACADM-rt-circRNA |
rt-circRNA-ACADM-RABGGTB-e7e2 |
0.0000059 |
-2.814912 |
down |
|
DoABR |
DoABR |
0.000000034 |
3.10297237 |
up |
|
DoABR-pre-mRNA |
ABR-pre-mRNA |
1.6E-09 |
5.44875714 |
up |
|
DoABR-sense-hsa_circ_0004931 |
hsa_circ_0004931 |
0.000000334 |
2.96418948 |
up |
|
DoABR-ChimeraRNA |
ChimeraRNA-ABR-NXN-e16e2 |
4.16E-08 |
4.49189189 |
up |
|
Annotation |
||||
|
DoGRegion |
DoGRegion |
HostGene |
Sense overlapping |
Anti-Sense overlapping |
|
chr12:2697950- |
82000 |
CACNA1C |
lncRNA ----- LINC02371-202 || LINC02371; |
lncRNA ----- ITFG2-AS1-201 || ITFG2-AS1 |
|
chr1:75763720- |
41366 |
ACADM |
CircRNA ----- hsa_circ_0012969(NM_004582 2-6/9) ; |
|
|
chr17:998017- |
5501 |
ABR |
ChimeraRNA || ABR(ENST00000302538) 1/23 || NXN(ENST00000336868) 2/8 ; |
CircRNA ----- hsa_circ_0001967(NM_013337 2-3/4) ; |
Transcript ID: DoG RNA的ID編號。
transcript_name:DoG RNA或相關轉錄本的名稱。
P-value:評估兩組DoG表達水平的差異是否具有統計學顯著性的p值。
|log2FC|:差異倍數作log2轉換的絕對值,表示实验组与对照组之间DoG表達水平的變化倍數。
Regulation:兩組比較的上調(up)或者下调(down)。
DoGRegion_Locus:DoG RNA所在的基因組位置,以染色体号和起始、终止位置表示。
DoGRegion_Length:DoG RNA的長度,以碱基数(nt)为单位。
HostGene:DoG RNA的來源基因名稱。
Sense overlapping DownstreamTranscripts:與DoG RNA在同方向上重疊的下游轉錄本。
Anti-Sense overlapping DownstreamTranscripts:與DoG RNA在反方向上重疊的下游轉錄本。
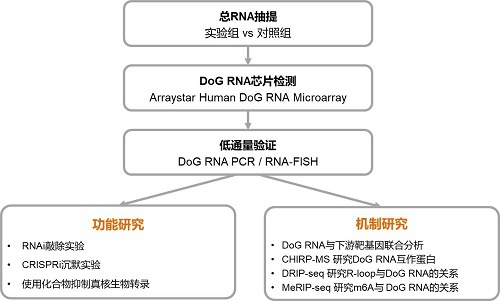
圖3. DoG RNA研究路線
參考文獻
1. Eaton JD et al: Xrn2 accelerates termination by RNA polymerase II, which is underpinned by CPSF73 activity. Genes Dev. 2018 Jan 15;32(2):127-139. PMID: 29432121; PMCID: PMC5830926.
2. Iwakiri J et al: Remarkable improvement in detection of readthrough downstream-of-gene transcripts by semi-extractable RNA-sequencing. RNA. 2023 Feb;29(2):170-177. PMID: 36384963; PMCID: PMC9891252.
3. Rutkowski AJ et al: Widespread disruption of host transcription termination in HSV-1 infection. Nat Commun. 2015 May 20;6:7126. PMID: 25989971; PMCID: PMC4441252.
4. Rosa-Mercado NA et al: Hyperosmotic stress alters the RNA polymerase II interactome and induces readthrough transcription despite widespread transcriptional repression. Mol Cell. 2021 Feb 4;81(3):502-513.e4. PMID: 33400923; PMCID: PMC7867636.
5. Cugusi S et al: Heat shock induces premature transcript termination and reconfigures the human transcriptome. Mol Cell. 2022 Apr 21;82(8):1573-1588.e10. PMID: 35114099; PMCID: PMC9098121.
6. Grosso AR et al: Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. Elife. 2015 Nov 17;4:e09214. PMID: 26575290; PMCID: PMC4744188.
7. Heinz S et al: Transcription Elongation Can Affect Genome 3D Structure. Cell. 2018 Sep 6;174(6):1522-1536.e22. PMID: 30146161; PMCID: PMC6130916.
8. Vilborg A et al: Widespread Inducible Transcription Downstream of Human Genes. Mol Cell. 2015 Aug 6;59(3):449-61. PMID: 26190259; PMCID: PMC4530028.
9. Hennig T et al: HSV-1-induced disruption of transcription termination resembles a cellular stress response but selectively increases chromatin accessibility downstream of genes. PLoS Pathog. 2018 Mar 26;14(3):e1006954. PMID: 29579120; PMCID: PMC5886697.
10. Dasilva LF et al: Integrator enforces the fidelity of transcriptional termination at protein-coding genes. Sci Adv. 2021 Nov 5;7(45):eabe3393. PMID: 34730992; PMCID: PMC8565846.
11. Roth SJ et al: ARTDeco: automatic readthrough transcription detection. BMC Bioinformatics. 2020 May 26;21(1):214. PMID: 32456667; PMCID: PMC7249449.
12. Wiesel Y et al: DoGFinder: a software for the discovery and quantification of readthrough transcripts from RNA-seq. BMC Genomics. 2018 Aug 8;19(1):597. PMID: 30089468; PMCID: PMC6083495.
13. Shah N et al: Tyrosine-1 of RNA Polymerase II CTD Controls Global Termination of Gene Transcription in Mammals. Mol Cell. 2018 Jan 4;69(1):48-61.e6. PMID: 29304333.
14. Glažar P et al: circBase: a database for circular RNAs. RNA. 2014 Nov;20(11):1666-70. PMID: 25234927; PMCID: PMC4201819.
15. Harrow J et al: GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012 Sep;22(9):1760-74. PMID: 22955987; PMCID: PMC3431492.
16. Birney E et al: An overview of Ensembl. Genome Res. 2004 May;14(5):925-8. PMID: 15078858; PMCID: PMC479121.
17. Grosso AR et al: Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. Elife. 2015 Nov 17;4:e09214. PMID: 26575290; PMCID: PMC4744188.
18. Vo JN et al: The Landscape of Circular RNA in Cancer. Cell. 2019 Feb 7;176(4):869-881.e13. PMID: 30735636; PMCID: PMC6601354.
19. Zhang Y et al. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016 Apr 19;15(3):611-624. PMID: 27068474.
20. Liang D et al. The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting. Mol Cell. 2017 Dec 7;68(5):940-954.e3. PMID: 29174924; PMCID: PMC5728686.
21. Varley KE et al. Recurrent read-through fusion transcripts in breast cancer. Breast Cancer Res Treat. 2014 Jul;146(2):287-97. PMID: 24929677; PMCID: PMC4085473.
22. Kim P et al. FusionGDB 2.0: fusion gene annotation update aided by deep learning. Nucleic Acids Res. 2021 Nov 10
23. Morgan M. et al. It's a DoG-eat-DoG world-altered transcriptional mechanisms drive downstream-of-gene (DoG) transcript production. Mol Cell. 2022; 82(11):1981-1991 [PMID:35487209]
24. Lai F. et al. Directed RNase H Cleavage of Nascent Transcripts Causes Transcription Termination. Mol Cell. 2020; 77(5):1032-1043.e4 [PMID:31924447]
25. RodrÃguez-Molina JB. et al. Knowing when to stop: Transcription termination on protein-coding genes by eukaryotic RNAPII. Mol Cell. 2023; 83(3):404-415 [PMID:36634677]
26. Hao JD et al: DDX21 mediates co-transcriptional RNA m(6)A modification to promote transcription termination and genome stability. Mol Cell 2024; 84(9):1711-1726 e1711.[PMID: 38569554]

